The matched sequences reported by search programs can be classified as true positives and false positives (the sequences missed by the program are the negatives). A true positive is a sequence that shares similarity with the query because both have evolved (diverged) from a common ancestral sequence, and is thus a true homolog. Similarity can also be sometimes attributed to evolutive convergence. A sequence is considered a false positive if the observed similarity is attributable to chance. It must be stressed that only biological arguments can enable one to decide whether a sequence should be regarded as a true or false positive. Nevertheless, a statistical analysis based on sound principles can help in the decision, because some matches are more likely to have been produced by chance than others (see statistical interpretation below). In addition we provide match status codes to help the biologist in interpreting the matches.
| ! | A strong match: it is very unlikely that this match is a false positive. | |||||||||||||||
| R |
Rescued match: despite the low score, it is considered to be a strong match. This concerns primarily domains known to be repeated and that are unlikely to occur as a single copy in a protein. |
|||||||||||||||
| ? | Questionable or weak match: determining the true or false negative status of this match requires additional biological evidences. | |||||||||||||||
| !! | Strong match for a family-specific motif: it is very unlikely that this match is a false positive, in addition it is very likely that this match belongs to the targeted sub-family. | |||||||||||||||
| ?! | Accepted match for a family-specific motif: it is very unlikely that this match is a false positive for the motif, but determining its family assignment requires additional biological evidences. | |||||||||||||||
| ?? | Questionable or weak match for a family-specific motif: determining the true or false negative status of this match requires additional biological evidences. | |||||||||||||||
| NA | Not Available. The above interpretation rules make no sense (e.g., for a low-complexity region). | |||||||||||||||
| Statistical Interpretation of a Match Score | ||||||||||||||||
| Statistics |
Similarity-search tools that use profiles or hidden Markov models (HMMs) as queries produce lists of matched sequences that are aligned with the query, either locally or semi-globally. Every match receives a numerical raw score , whose value is guaranteed to meet some local-maximum criterion. Only matches with scores greater than some threshold are usually reported, and every profile or HMM possesses its own score thresholds for reporting a match. To make the interpretation of the match scores a little easier, one generally attempts to "rescale" the score onto a new scale which is common to all predictors and which possesses a well-defined statistical meaning. Two such scales are the E-value of Pfam HMMs and the normalized score of Prosite profiles. How the rescaling is actually computed is out of the scope of this page. |
|||||||||||||||
| E-value |
The E-value is the number of matches with a score equal to or greater than the observed score that are expected to occur by chance. In other words, the E-value provides an estimation of the number of false positives. The E-value depends on the size of the database searched, as the number of false positives expected to be above a given score threshold usually increases proportionately with the size of the database. The total number of sequences and the total number of residues are the most frequently used measures for the database size. |
|||||||||||||||
| Normalized score |
The Prosite profiles report normalized scores instead of E-values, which are defined as the base 10 logarithm of the size (in residues) of the database in which one false positive match is expected to occur by chance. The normalized score is independent of the size of the databases searched. The so-called bit scores reported by other database-search programs have a distinct meaning but are also independent of the size of the database searched. For a given database size of DB_size residues, the normalized score N_score and the E-value are easily interconvertible:
N_Score =
The following table gives some examples of conversions. The calculations were made for release 34 of SwissProt of october 1996 which contained 21'210'388 = 107.33 residues (in 59'021 entries) and for all the sequences found in the Hits database in June 2001 which amount to a total of 408'011'338 = 108.61 residues (in 1'818'627 entries).log10 DB_size -log10 E-valueor E-value = DB_size*10-N_Score
|
|||||||||||||||
| How to Reach a Decision about Match Significance | ||||||||||||||||
The introduction of the E-value and normalized scores greatly facilitates the interpretation of the match raw scores. To go one step further, one would like to reach a decision about the final status of a match. Is a given match to be regarded with some confidence, or is it questionable, i.e. a weak match? Indeed several cutoffs are supplied with Prosite profiles and Pfam HMMs just for that purpose. The idealized figure below illustrates the problem of defining these cutoffs. Note that the complete list of homologs is a priori unknown in most real situations, constituting a serious complication.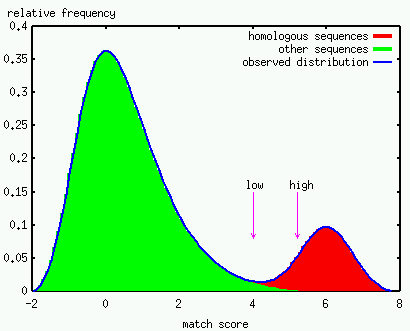
No false positive matches should be reported when considering the particular task of automated annotation of protein sequences. This dictates the use of cutoffs placed at a sufficiently high score, just to be on the safe side; this might correspond to the threshold indicated by the "high" arrow in the above figure. In principle, all the matches tagged with an "!" or "!!" in the output of the Motif Scan Server should belong to this category. The same cutoffs are used by the InterPro team for the Prosite profiles and the Pfam HMMs, even if the scores of the matches are actually not reported by the InterProScan server (June 2001). Gene discovery and the detection of remote homologs are other tasks where profile and HMMs have proven to be successful. In this perspective, inspection of the matches in the twilight zone where true and false positives co-exist often yielded the most promising results. This would include the scores located between the "low" and "high" arrows on the above idealized figure. These weak or questionable matches are tagged with a "?" or "??" in the output of the Motif Scan Server and further investigation is necessary before being reported elsewhere. Note also that (i) the normalized score is the most helpful to evaluate matches in this zone; (ii) the low score threshold is arbitrarily set for most predictors; (iii) the use of meta-motifs is usually an efficient and elegant manner to further evaluate weak matches. |
||||||||||||||||